CD Laboratory for Applied metabolomics
Head of research unit

Commercial Partner
Duration
Thematic Cluster
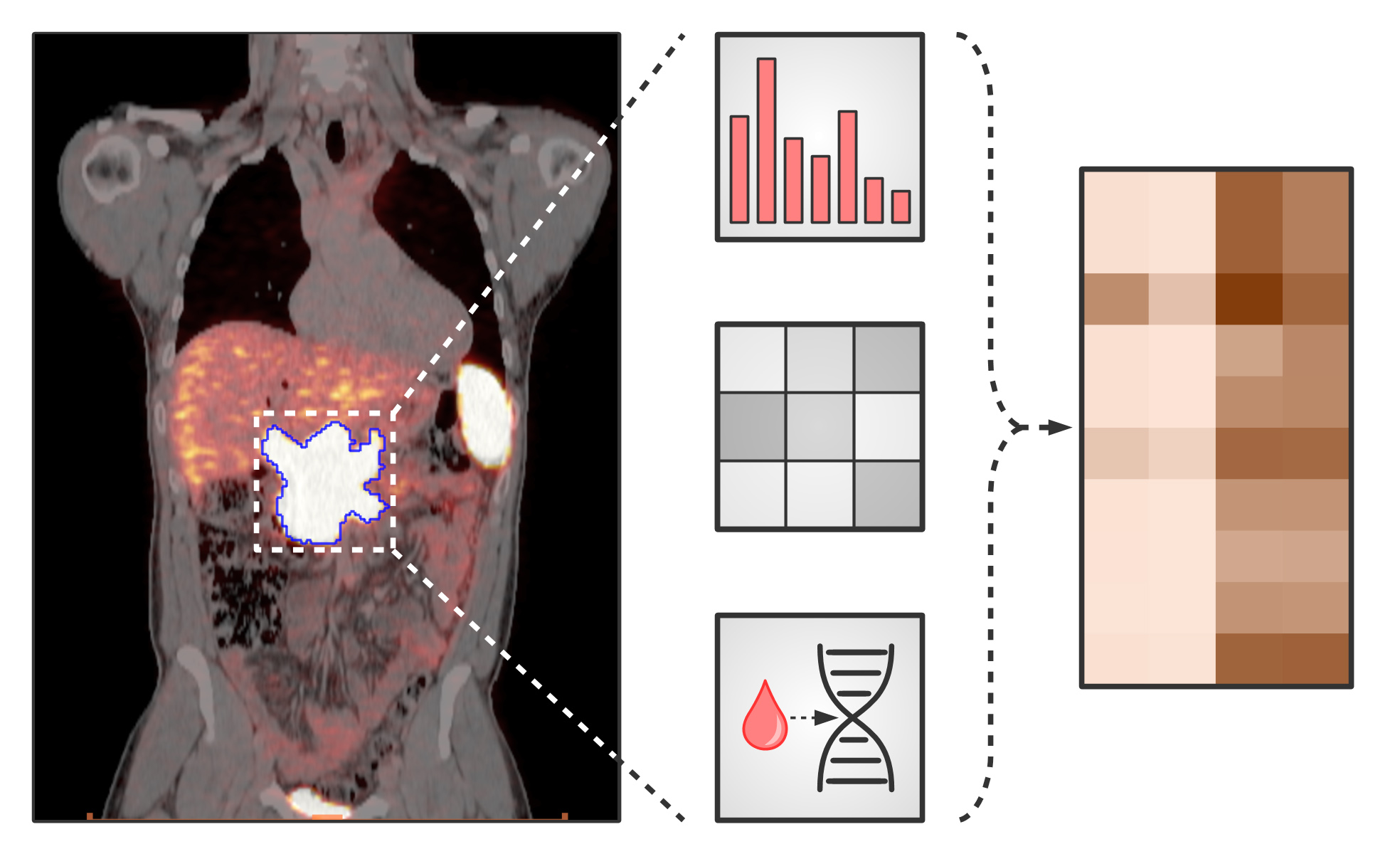
This CD Laboratory is researching ways to better characterise tumours using non-invasive imaging techniques. This is important as tumours are constantly changing due to mutations. This should make it possible to personalise treatment and monitor its success on an ongoing basis.
New, targeted therapies that attack gene mutations or specific receptors also require more refined molecular tumour characterisation. The biology of tumour cells is constantly changing due to new mutations. This process occurs extremely quickly in some types of tumour. Therefore, a non-invasive diagnostic method that can map these changes is of eminent importance. With the growing number of targeted therapeutics that enable patients to receive optimal, personalised therapy, the need for non-invasive prognostic biomarkers to identify which person will respond to which therapy is also increasing. Positron emission tomography (PET) is well established in oncology and available in many hospitals, but only a fraction of its potential is currently being utilised. Genetic changes in the tumour produce characteristic metabolic patterns that can be detected using PET.
The CD Laboratory for non-invasive tumour characterisation aims to validate innovative PET imaging using classical methods. The PET data obtained will be combined with histopathological examinations, analyses of therapy and prognosis-relevant genetic mutations, as well as clinical data on survival and therapy response. The result should be a non-invasive ""in vivo pathology"" that leads to an individualised therapy algorithm and can continuously monitor its success. To achieve this complex goal, a supervised machine learning (ML) algorithm will be used. This novel method is intended to identify specific PET patterns caused by mutations in the tumour and also provide information on the response to therapy.
The results are validated with the help of preclinical mouse models. Using CRISPR/Cas9, genetic mutations are induced in xenografts, which in this case are mice to which human tumour cells have been transferred, and then examined to see whether the PET patterns found in patients can be reproduced in the mouse model. In addition, the imaging approach is prospectively validated with liquid biopsies, i.e. examinations for tumour components and tumour markers in the blood. Blood samples are taken shortly before PET imaging and clinical data such as survival and response to therapy are collected so that they can be correlated with the PET patterns. The overall goal is to develop an integrative diagnostic algorithm to non-invasively determine and monitor the tumour biology and thus the best possible initial therapy.
In order to develop the pipeline described, three tumour entities were selected that pose different challenges for the method to be investigated: colorectal carcinomas and diffuse large B-cell lymphomas - both investigated using F18-2 fluorodeoxyglucose (FDG) PET - and prostate carcinomas - investigated using prostate-specific membrane antigen (PSMA) PET. In the further course, the study will be extended to breast carcinomas and non-small cell lung carcinomas.
Christian Doppler Forschungsgesellschaft
Boltzmanngasse 20/1/3 | 1090 Wien | Tel: +43 1 5042205 | Fax: +43 1 5042205-20 | office@cdg.ac.at


